Design and Selection of Antisense Oligonucleotides Against miRNA
To design antisense oligonucleotides (ASOs) targeting a specific microRNA (miRNA), a multi-step computational approach was employed. The strategy involved sequence-based selection, secondary structure prediction, molecular docking, and statistical validation of RNA-RNA interactions. The following computational tools and techniques were used:
- miRNA Template Selection and Analysis: The miRNA sequence targeted for inhibition was retrieved from the miRBase database. This sequence served as the template for antisense oligonucleotide design. The selected miRNA was known to be associated with a specific disease phenotype, and its role in gene silencing was experimentally validated in previous studies.
- Secondary Structure Prediction using UNAfold: The secondary structure of the miRNA was predicted using the UNAfold software package (Markham & Zuker, 2008). UNAfold uses thermodynamic principles to predict RNA folding and secondary structures by calculating free energy changes during folding. The folding of the miRNA sequence was simulated at a physiological temperature (37°C) and ionic conditions (1 M Na+ concentration). This step was critical for identifying potential binding sites accessible for antisense oligonucleotides (ASOs). Secondary structures with the lowest Gibbs free energy (ΔG) were prioritized as they represent the most stable conformations of the miRNA. Binding site accessibility was further analyzed by evaluating single-stranded regions that are suitable for ASO targeting.
- Tertiary Structure Modeling using RNA Composer: To gain insights into the 3D conformation of the miRNA, the tertiary structure was modeled using RNA Composer (Popenda et al., 2012; Sarzynska et al., 2023), a web server for automated RNA 3D structure prediction based on secondary structure information. The RNA Composer utilizes fragment-based approaches and knowledge-based algorithms to construct 3D models from predicted or known secondary structures. The predicted 3D structure of the miRNA was used to identify surface-exposed regions, loops, and hairpins that are potentially accessible to antisense oligonucleotide binding.
- RNA-RNA Docking using HNADOCK: After predicting the tertiary structure of the miRNA, molecular docking was performed to simulate RNA-RNA interactions between the miRNA and candidate ASOs using HNADOCK (He et al., 2019). HNADOCK is a molecular docking tool specifically designed for studying nucleic acid complexes, leveraging an RNA-specific force field and docking algorithms. Candidate ASOs were selected based on their complementarity to the miRNA target and were docked onto the 3D structure of the miRNA to evaluate binding affinity and complex stability. Docking was performed with a flexible RNA backbone for both the miRNA and the ASO to account for conformational changes during binding. The resulting RNA-RNA complexes were ranked based on their predicted binding energies, with the top-scoring ASO-miRNA complexes chosen for further analysis.
Elucidation of RNA-RNA Complexes using Kullback-Leibler (KL) Distance
To quantitatively evaluate the similarity and divergence between the predicted RNA-RNA complexes, Kullback-Leibler (KL) distance was employed. The KL distance, also known as relative entropy, is a statistical measure that quantifies the difference between two probability distributions. In the context of RNA-RNA interactions, the KL distance was used to compare the base-pairing probability distributions of the free miRNA and its complexes with various ASOs. The base-pairing probabilities for the free miRNA and the ASO-bound miRNA were obtained from the UNAfold partition function output.
The KL distance between the free miRNA and the ASO-miRNA complex was calculated using the following formula:
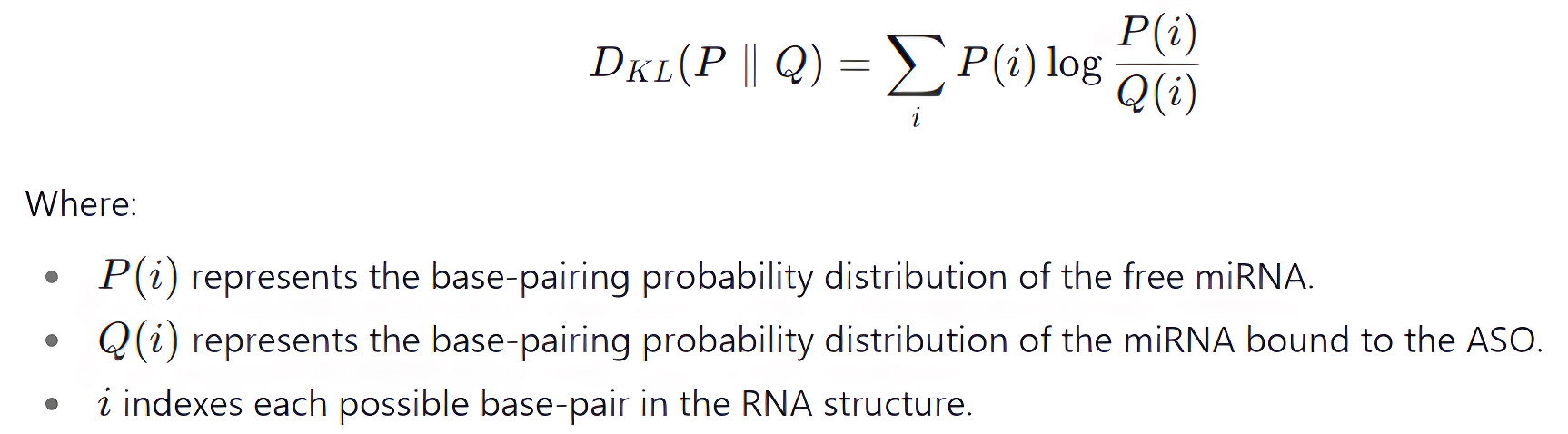
A higher KL distance indicates greater divergence in the base-pairing patterns between the free and bound miRNA, suggesting significant structural changes upon ASO binding. The ASO-miRNA complexes with lower KL distances were prioritized, as they indicated minimal perturbation of the miRNA's natural folding, which may correlate with improved target specificity and fewer off-target effects.
Statistical Validation of RNA-RNA Interactions
To statistically validate the docking results and KL distance calculations, several measures were taken:
- Binding Energy Correlation: The predicted binding energies from HNADOCK were correlated with the KL distances to assess whether structural changes upon binding correlated with stronger interactions. Pearson's correlation coefficient (r) was calculated between binding energies and KL distances to determine the statistical significance of the relationship between binding strength and structural divergence.
- Monte Carlo Simulations: Monte Carlo simulations were performed to assess the robustness of the RNA-RNA docking and KL distance results. Multiple rounds of simulations were conducted by randomly perturbing the RNA sequences to generate alternative structures. The consistency of the docking and KL distance outcomes across these simulations was evaluated to confirm the reliability of the results.
- p-value Calculation: The statistical significance of the differences in KL distances between the free miRNA and the ASO-bound complexes was determined by calculating p-values using a non-parametric bootstrapping approach. A p-value threshold of 0.05 was used to determine the significance of the ASO's effect on miRNA structure and binding.
In summary, this computational workflow integrates thermodynamic, structural, and statistical analyses to design antisense oligonucleotides with high specificity and efficacy against miRNA targets. The use of UNAfold, RNA Composer, and HNADOCK allowed for comprehensive prediction of miRNA structures and interactions, while the Kullback-Leibler distance provided a robust method for quantitatively assessing RNA-RNA complex formation.